
by Library News
This is a guest post by Dr. Cara Boutte about her participation in the Libraries' Experiential Learning Faculty Fellow program (see Funding Opportunities).
Everything interesting in Biology happens in 3-dimensional space, but we are typically restricted to 2-dimensional teaching modalities: images in PowerPoint slides, papers, and textbooks. This is not such a great barrier to teaching organismal biology and anatomy, as students can see and interact with the subjects in three dimensions in real life, due to their size. However, students cannot typically interact with the molecules that make up all living things in three dimensions, and this makes it more difficult for them to build a mental model of key concepts in molecular biology.
In BIOL_3306, Bacterial Physiology and Antibiotics, I use the primary literature to help teach key concepts in Bacterial Physiology, with a focus on the functioning of key antibiotic targets. Antibiotics are small molecules that bind and inhibit the function of proteins that are essential for the survival of bacteria. Proteins are 3-dimensional molecules, and their 3D shape is critical for their function: antibiotics bind to specific pockets within the 3D molecules. When I have taught this class previously, I show students 2-dimensional pictures of proteins and antibiotics, but I think they have typically struggled to grasp both how proteins work to perform their essential functions, and how antibiotic inhibition works.
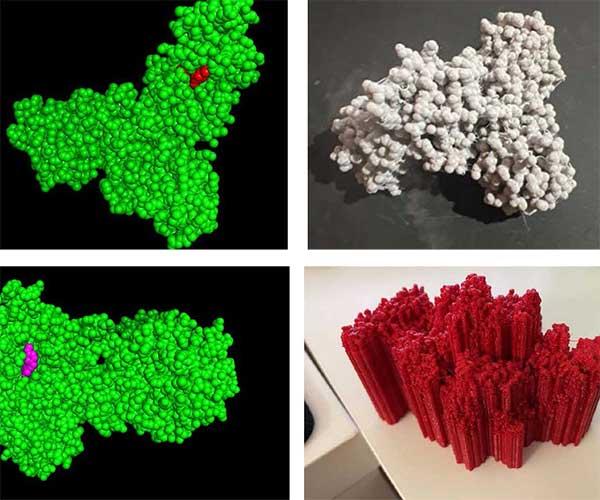
This year, I used the ELFF fellowship to introduce a module into the class where the students learn to use software that allows them to visualize the 3-dimensional structure of proteins, and to see how tightly the antibiotic molecules fit within specifically-shaped pockets within the protein structure. Each student then picked a protein-antibiotic structure, and we had a class where we went to the library and worked with the FabLab staff to prepare the digital files for 3D printing. Each student printed their protein-antibiotic structure and did an assignment where they answered questions about it.
This assignment had students download 3D protein models from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB). Next, they import their downloaded files into Pymol, a protein visualization platform. In Pymol, they view the proteins, switch from the structural view to the spherical view, highlight the segments that represent the antibiotic attached to the protein, and save the files as .stl files for printing. Most models can be sent directly to KISSlicer, an application used to transform the models into G-Code, the software language used by our 3D printers. Models over a certain size threshold, and those with antibiotics deeply imbedded in the protein (requiring a two-color printing process) would first need to be resized and color-separated in Meshmixer before going to KISSlicer.
I think this activity helped the students to understand proteins as 3-dimensional objects, and I think this increased comprehension of some of the key concepts in the class for some students. However, a limitation of the 3D prints was that we were unable to use 2-colors, as the multi-color 3D printer was not functioning properly. So, in the final 3D printed structures, it was not possible to distinguish the protein from the antibiotic. This limited the helpfulness of this activity in promoting understanding of antibiotic function.
I really appreciate the Library and FabLab staff in helping introduce this new activity to my class. I hope that we can get 2-color 3D printing working, which would allow me to continue using this learning activity in my class.
Add new comment